5. Fire up R
If you're following along at home jump to installation.
> # check it out we can write comment characters in R too
> # I'm loading the file with a header row, with the column
> # separator set to be a tab, and with quotes as quotes.
> counties <- read.delim("/path/to/your/file/county_demographics.txt",header=TRUE, sep="\t", quote='"')
> # What are the column names?
> colnames(counties)
[1] "state" "co" "pop" "median_hh_income" "ba_plus" "white_pcnt" "black_pcnt"
[8] "hispanic_pcnt" "pov_pcnt"
> # This is a bit odd, but this data is now stored in R as a 'data frame'. Don't worry about this for the moment.
> # Here's how we access data in a data frame:
> counties$state
Accessing the data like that prints it all out. We can summarize it too:
> summary(counties$state)
Ala. Alaska Ariz. Ark. Calif. Colo. Conn. D.C. Del. Fla Ga. Hawaii Idaho Ill. Ind. Iowa Kansas Ky.
67 27 15 75 58 63 8 1 3 67 159 5 44 102 92 99 105 120
La. Maine Mass. Md. Mich. Minn. Miss. Mo. Mont. N. Dak. N. Mex. N.C. N.H. N.J. N.Y. Neb. Nev. Ohio
64 16 14 24 83 87 82 115 56 53 33 100 10 21 62 93 17 88
Okla. Ore. Pa. R.I. S.C. S.D. Tenn. Texas Utah Va. Vt. W. Va. Wash. Wis. Wyo.
77 36 67 5 46 66 95 254 29 134 14 55 39 72 23
We can also summarize the whole data frame:
> summary(counties)
state co pop median_hh_income ba_plus white_pcnt
Texas : 254 Washington Co.: 30 Min. : 78 Min. : 18869 Min. : 5.00 Min. : 6.00
Ga. : 159 Jefferson Co. : 25 1st Qu.: 11011 1st Qu.: 35979 1st Qu.:13.00 1st Qu.: 77.00
Va. : 134 Franklin Co. : 24 Median : 25436 Median : 41652 Median :17.00 Median : 90.00
Ky. : 120 Jackson Co. : 23 Mean : 96077 Mean : 43435 Mean :18.68 Mean : 83.92
Mo. : 115 Lincoln Co. : 23 3rd Qu.: 65524 3rd Qu.: 48285 3rd Qu.:22.00 3rd Qu.: 96.00
Kansas : 105 Madison Co. : 19 Max. :9785295 Max. :113313 Max. :69.00 Max. :100.00
(Other):2253 (Other) :2996 NA's : 3 NA's : 3 NA's : 3.00 NA's : 3.00
black_pcnt hispanic_pcnt pov_pcnt
Min. : 0.000 Min. : 0.000 Min. : 1.00
1st Qu.: 1.000 1st Qu.: 1.000 1st Qu.:11.00
Median : 3.000 Median : 3.000 Median :14.00
Mean : 9.618 Mean : 7.623 Mean :15.27
3rd Qu.: 12.000 3rd Qu.: 7.000 3rd Qu.:19.00
Max. : 87.000 Max. :99.000 Max. :47.00
NA's :252.000 NA's :46.000 NA's : 5.00
Lets take a minute to look at that summary. See how it correctly identifies text and numbers. That's good.
See the NA's. That's how many values are missing. There could be missing values in the text columns too, it just turns out that in this data set they aren't missing. Also note how the data is summarized for numerical quantities: Minimum, 1st quartile, median, mean, quartile, 3rd quartile, max.
¡ ¡ Malformed data files with unclosed quotes will confuse R--typically resulting in fewer lines than expected. So it's a good idea to check that all your rows got there.
> # how many rows are in the data altogether?
> nrow(counties)
[1] 3140
> # lets copy the counties
> counties2 <- counties
We can also assign new columns in a data frame with a similar syntax.
> # lets change units in income. What did I say income was again?
> colnames(counties2)
[1] "state" "co" "pop" "median_hh_income" "ba_plus" "white_pcnt"
[7] "black_pcnt" "hispanic_pcnt" "pov_pcnt"
> # right, it's median_hh_income, (Good ole B19013 for census peeps)
> counties2$income = counties2$median_hh_income/1000
> # did it work?
> counties2$income[0]
numeric(0)
# Whoa--that's not what you might have been expecting
# The zeroeth entry tells us the data type.
# In other words, the columns that are the data are 1-indexed
# Let's see the first row:
> counties2$income[1]
[1] 51.463
> counties$median_hh_income[1]
[1] 51463
Relationships between the data
9. Look at the data more
R's plot command is pretty robust. Lets get to it.
> plot(counties$median_hh_income, counties$ba_plus)
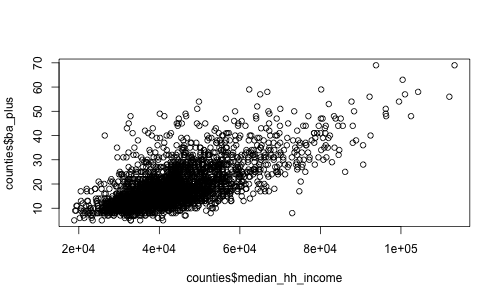
So there's obviously a relationship there. How much?
> cor(counties$median_hh_income, counties$ba_plus)
[1] NA
> # We can't correlate because there are null values.
> cor(counties$median_hh_income, counties$ba_plus, use="complete.obs")
[1] 0.6763103
In the above, we're just saying use only the complete observations. That could be dangerous.
Also note--the default is pearson correlation. But sometimes it's useful to use spearman (rank)
> cor(counties$median_hh_income, counties$ba_plus, use="complete.obs", method='pearson')
[1] 0.6763103
> cor(counties$median_hh_income, counties$ba_plus, use="complete.obs", method='spearman')
[1] 0.6236564
What about the uncertainty of the correlation?
> cor.test(counties$median_hh_income, counties$ba_plus, use="complete.obs")
Pearson's product-moment correlation
data: counties$median_hh_income and counties$ba_plus
t = 51.4071, df = 3135, p-value < 2.2e-16
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
0.6568609 0.6948604
sample estimates:
cor
0.6763103
>
But we're only examining one relationship this way. Lets do all of them.
I'm gonna transform it into a matrix. Don't ask why, I'm not really sure.
> county_matrix <- data.matrix(counties)
> cor(county_matrix, use="complete.obs")
state co pop median_hh_income ba_plus
state 1.00000000 0.013705958 -0.05401844 0.03493939 0.035086788
co 0.01370596 1.000000000 0.01460693 0.02956972 0.004772263
pop -0.05401844 0.014606927 1.00000000 0.26945683 0.325710503
median_hh_income 0.03493939 0.029569722 0.26945683 1.00000000 0.685827752
ba_plus 0.03508679 0.004772263 0.32571050 0.68582775 1.000000000
white_pcnt 0.12284483 0.002043147 -0.17966726 0.13377545 0.013097471
black_pcnt -0.12748272 -0.008496116 0.06352790 -0.23045513 -0.097418256
hispanic_pcnt 0.08728682 0.028717294 0.18910339 0.01684250 0.020222111
pov_pcnt -0.06326769 -0.014163910 -0.13208919 -0.79398962 -0.432999042
white_pcnt black_pcnt hispanic_pcnt pov_pcnt
state 0.122844834 -0.127482722 0.08728682 -0.06326769
co 0.002043147 -0.008496116 0.02871729 -0.01416391
pop -0.179667258 0.063527896 0.18910339 -0.13208919
median_hh_income 0.133775455 -0.230455130 0.01684250 -0.79398962
ba_plus 0.013097471 -0.097418256 0.02022211 -0.43299904
white_pcnt 1.000000000 -0.832365355 -0.15548590 -0.41024198
black_pcnt -0.832365355 1.000000000 -0.11994898 0.42466780
hispanic_pcnt -0.155485898 -0.119948976 1.00000000 0.09953580
pov_pcnt -0.410241980 0.424667802 0.09953580 1.00000000
That's hard to read, lets write it out to a table in the filesystem:
> cor_found <- cor(county_matrix, use="complete.obs")
> write.table( cor_found, file="correlations.txt", sep="|", eol="\n", row.names=TRUE)
| state | co | pop | income | ba_plus | white_pcnt | black_pcnt | hispanic_pcnt | pov_pcnt |
|---|
| state | 1.00 | 0.01 | -0.05 | 0.03 | 0.04 | 0.12 | -0.13 | 0.09 | -0.06 |
| co | 0.01 | 1.00 | 0.01 | 0.03 | 0.00 | 0.00 | -0.01 | 0.03 | -0.01 |
| pop | -0.05 | 0.01 | 1.00 | 0.27 | 0.33 | -0.18 | 0.06 | 0.19 | -0.13 |
| income | 0.03 | 0.03 | 0.27 | 1.00 | 0.69 | 0.13 | -0.23 | 0.02 | -0.79 |
| ba_plus | 0.04 | 0.00 | 0.33 | 0.69 | 1.00 | 0.01 | -0.10 | 0.02 | -0.43 |
| white_pcnt | 0.12 | 0.00 | -0.18 | 0.13 | 0.01 | 1.00 | -0.83 | -0.16 | -0.41 |
| black_pcnt | -0.13 | -0.01 | 0.06 | -0.23 | -0.10 | -0.83 | 1.00 | -0.12 | 0.42 |
| hispanic_pcnt | 0.09 | 0.03 | 0.19 | 0.02 | 0.02 | -0.16 | -0.12 | 1.00 | 0.10 |
| pov_pcnt | -0.06 | -0.01 | -0.13 | -0.79 | -0.43 | -0.41 | 0.42 | 0.10 | 1.00 |
10. Pearson correlation is for *linear* correlation
Shamelessly ripped off from wikipedia
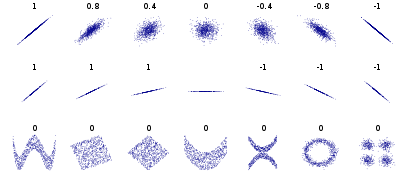
If you're looking for *non-linear* relationships use spearman correlation...
Done!
But if we're not out of time yet, here's more
11. Adding libraries
All kinds of good stuff is put out by random people (typically people who know way more about it than I). Stuff is different on windows. Here's how it works on Mac, roughly:
- "Packages & Data" > "R Package Manager" lots of stuff ships with R, you might just have to load it.
on macs, click load, then, to load it at the console> library(ggplot2)
- Can load the data via repository; in macs: "Packages & Data" > "R Package Installer"
I skipped this because it varies with platform. It's pretty painless on my rig, just remember to click the 'add dependencies' button, which oddly isn't checked by default.
12. Getting help
There's a system help. It's not incredibly helpful for beginners (though it's invaluable if you really want to know how something works). You can get to it with windows. In general, typing '?command' at the console *should* launch it, but in the labs it doesn't. I could make it work by cutting a pasting the local help url--once it's started in the labs--into a browser bar.
http://127.0.0.1:10788/doc/html/index.html This might work in the labs (?)
http://127.0.0.1:10788/library/base/html/getwd.html
- "Packages & Data" > "R Package Manager" lots of stuff ships with R, you might just have to load it.
on macs, click load, then, to load it at the console> library(ggplot2)
- Can load the data via repository; in macs: "Packages & Data" > "R Package Installer"
- The reality is--it's usually more helpful to google for examples; try stackoverflow etc. Also, the name 'R' is totally unhelpful for googling purposes. Which is a funny problem. Try 'r statistics' + your help term.
- I mentioned this before, but you can see the source code of a function by just typing it at the console window without parentheses. Not sure that's actually helpful, but...
13. Working example: dates, ggplot2, etc.
Here's a data file, call it faceted.txt
a <- read.delim("/Users/jacobfenton/IRW/banks_small_biz/parse_custom/custom_read.csv",header=TRUE, sep=",", quote='"')
14. Installation
There are installers for Mac and for windows. Installing with homebrew to use at the command line, for example, seems to make it harder to do thing like load and install packages from remote libraries.